Metagenomics sequencing offers a direct, culture-free way to explore the full complexity of microbial communities in clinical and environmental samples. At Source Genomics, we work closely with researchers to turn this powerful approach into clear, high-resolution insights that reveal both the structure and function of the microbial world. With this level of insight, you can investigate biodiversity, track microbial shifts, and identify functional markers through a solution that’s both precise and scalable, built to support the specific goals of your research.
Metagenomics Sequencing Services
Our comprehensive platform enables detailed microbial analysis at every level:
16S rRNA Sequencing
Provides accurate identification of bacterial and archaeal populations.
18S rRNA Sequencing
Targets eukaryotic microbes such as protists and fungi for broad taxonomic profiling.
ITS Sequencing
Offers high-resolution characterisation of fungal communities.
DNA Extraction Services
Delivers high-quality DNA suitable for sequencing from diverse and complex sample types.
Custom Amplicon Design
Facilitates tailored PCR workflows to match specific research requirements.
Sequencing & Bioinformatics
Generates 300 bp paired-end reads with full taxonomic analysis and comprehensive reporting.
The metagenomics sequencing services from Source Genomics deliver the depth and precision needed to study complex microbial communities. They support research in microbiome analysis, species identification, and diversity profiling, as well as a wide range of other microbial investigations. Clients can apply them to evaluate soil and water quality, explore gut microbiomes, study fungal populations, and detect health-related biomarkers. With high-resolution, scalable data, these studies become more focused, reproducible, and impactful.
Contact us to discuss your project today. Our team is committed to the highest quality standards and fast turnaround times, ensuring your research needs are always our top priority.
Designed for Depth
Source Genomics delivers more than sequencing services. We provide a collaborative, research-driven approach that ensures every dataset is tailored to your scientific objectives. Our process is built on transparency, technical precision, and expert guidance at every stage. With ISO 9001:2015-certified protocols and a dedicated support team, your results will meet the highest standards for scientific integrity and reliability.
The Metagenomics Sequencing Process
Initial Consultation
Define your project goals and sample types with our team to determine the most effective approach.
Sample QC
Evaluate the integrity and suitability of your samples before sequencing begins.
Library Preparation
Prepare samples using a robust two-step PCR protocol with dual indexing to ensure clean, accurate reads.
Sequencing
Perform high-throughput sequencing using 300 bp paired-end reads on the Illumina MiSeq platform.
Bioinformatics (Optional)
Apply advanced tools for taxonomic classification, OTU clustering, and custom data visualisation.
Data Delivery
Receive results securely via FTP or physical drive, ready for immediate analysis.
Benefits
Choosing Source Genomics for your metagenomics sequencing means gaining access to reliable data, expert support, and workflows designed to maximise scientific value.
Overview
Sequencing package details
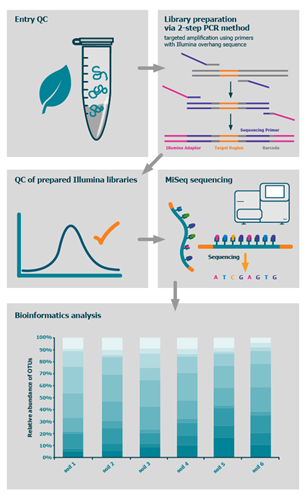
The targeted metagenomics sequencing package utilises a 2-step PCR library preparation approach to amplify your choice of 16S, 18S or ITS regions and incorporate unique dual indexes and Illumina adapters into the library for sequencing. Samples are sequenced using the Illumina MiSeq instrument in the 300bp paired-end read mode to a guaranteed minimum of 40,000 read pairs per sample.
By using a 2-step PCR approach, the package can also be easily adapted for any amplicon with complimentary overhangs to undergo the 2nd index PCR step. Please consult our Genomics team for more information.
Our standard targeted metagenomics package includes:
- Sample entry QC
- Library preparation via 2-step PCR protocol
- Illumina 300bp PE sequencing, guaranteed >40,000 read pairs per sample
- Return of FastQ files via secure FTP or HDD
Optional extras:
- DNA Extraction
- Bioinformatics analysis
Custom Target
Samples for 2nd index PCR only should be prepared using primers with overhangs. Primers should be designed as below and resulting PCR product should form a discrete band on a gel with length of 450-550bp.
Forward primer: 5’ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG‐[locus-primer]
Reverse primer: 5’ GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG‐[locus-primer]
Samples will then undergo 2nd index PCR step and subsequent sequencing to 40,000 read pairs.
BioInformatics Taxonomical Classification
Taxonomic classification of mixed samples to provide a profile of microbial diversity
By sequencing conserved regions of ribosomal RNA – 16S for prokaryotic and 18S for eurkaryotic, or the ITS region for fungal samples – taxonomic classification can be achieved. The analysis involves trimming raw sequencing reads, picking operational taxonomic units (OTUs), matching these against a database of choice, such as GreenGenes or Silva, to obtain a break-down for each sample at each of 7 levels: kingdom, phylum, class, order, family, genus and species. You will receive raw sequencing files (fastq), text files detailing OTUs and taxonomic classifications as well as bar charts for each sample. A bioinformatics report will provide further detail about the tools used and a summary of results.
The process
How to order
Contact us today and one of our skilled account managers will be in touch with a free consultation including further information and pricing details.
Payment options:
Payment can be made by credit card or purchase order number.
FAQ's:
View our frequently asked questions for more information.
Sample requirements
Targeted metagenomics package requires 100ng of gDNA (OD260/280 ratio ranging from 1.8-2) at a minimum concentration of 10 ng/μl and in a minimum volume of 10 μl. Click here for more details.
Sequencing Platform
|
Platform Type |
Illumina NovaSeq 6000 |
|
Read Length |
Paired-end 250 bp |
|
Recommended Sequencing Depth |
30K/50K/100K reads |
Ready to Begin?
Start your next microbial research project with sequencing you can trust. Source Genomics delivers scientifically robust results, streamlined workflows, and expert-led support that makes complex data easier to understand and act on. Get in touch today and see how we can support your work.
Contact us today and one of our skilled account managers will be in touch with a free consultation including further information and pricing details.
Next Generation Sequencing
Source BioScience is one of Europe’s leading providers of commercial sequencing, offering Next Generation Sequencing services from our ISO accredited laboratories. We offer NGS services on the most prominent platforms including Illumina’s NovaSeq, NextSeq and MiSeq.