Whole exome sequencing (WES) focuses on the protein-coding regions of the genome, which make up about 1-2% of the entire genome. By only sequencing these protein-coding regions, WES sequencing is much more cost-effective compared to whole genome sequencing (WGS). Whole exome sequencing is a powerful method for identifying genetic variants linked to various genetic diseases.
Source Genomics provides a variety of customisable packages to meet your specific research needs. Using the Illumina DNA Prep protocol for enrichment library preparation, we offer multiple capture panels to best match your project requirements, ensuring high-quality sequencing reads and optimal sequencing coverage.
Our primary capture panel is the Agilent SureSelect Human All Exon V7, but we also use other panels such as the Twist All Exon and the IDT Exome Research panel. Additionally, we can accommodate other capture panels, including custom panels designed specifically for your project. Our exome sequencing WES solutions are tailored to deliver reliable data for your whole exome sequencing analysis.
Contact us to discuss your project today. Our team is committed to the highest quality standards and fast turnaround times, ensuring your research needs are always our top priority.
Overview
Sequencing package details
Our standard sequencing package provides 100x raw coverage, using the SureSelect Human All Exon panel. This typically results in a mean coverage of 27x for over 95% of the targeted bases from approximately 5Gb of data, often exceeding Agilent's specifications.
Our standard package offering includes:
- Exome enrichment and library preparation
- Illumina 150bp PE sequencing – 100x raw coverage – 5Gb
- Return of raw FastQ files via secure FTP or HDD
Additional services available upon request:
- Increased sequencing depth
- DNA extraction
- Bioinformatics analysis
We also offer advanced BioIT variant analysis packages, as well as fully customised BioIT solutions to analyse your sequence data, identify genetic variants, and ensure high-quality output.
The process
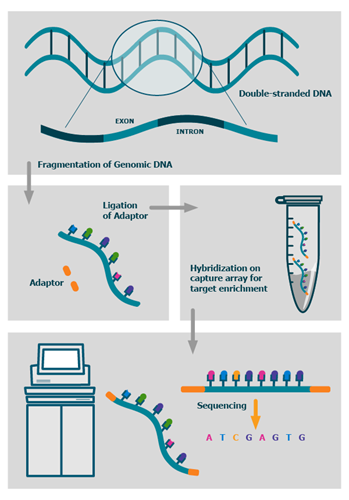
We also offer advanced BioIT variant analysis packages as well as fully bespoke BioIT solutions.
BioInformatics Variant Calling
Germline:
We investigate germline single nucleotide polymorphisms (SNPs), insertions, and deletions (INDELs) present in whole genome or exome data.
This analysis uses next-generation sequencing (NGS) data to identify germline mutations by comparing your data to a reference genome. Raw reads are trimmed, aligned, and deduplicated, followed by variant calling, recalibration, filtering, and annotation using public databases like dbSNP and ClinVar. You will receive raw (FastQ) and aligned (BAM) files for each sample, as well as variant call format (VCF) files listing all identified variants. Additionally, a bioinformatics report will provide detailed information on the tools used, alignment metrics, and variant quality statistics.
We also offer access to an interactive online viewing tool (Relyter) for enhanced data analysis of your exome sequencing WES results.
Somatic Variant Analysis:
We also perform analysis to identify and quantify somatic single nucleotide variants (SNVs), insertions, and deletions (INDELs) in cancer samples.
This process involves using NGS data to compare cancer samples against either a reference genome or a matched healthy sample. Raw reads are trimmed, aligned, and deduplicated, and somatic variants are identified through local de-novo assembly of haplotypes in active regions. Regions showing potential somatic variation are reassembled to create candidate variant haplotypes. Additional steps include contamination estimation, detection of orientation bias, and filtering, followed by functional annotation using databases like GENCODE and dbSNP. You will receive raw (FastQ) and aligned (BAM) files for each sample, as well as VCF files of all variants identified. A bioinformatics report will provide detailed information about the tools used, alignment metrics, and variant quality statistics.
How to order
Contact us today, and one of our skilled account managers will reach out to provide a free consultation, including more information and pricing details.
Payment options:
We accept payments via credit card or purchase order number.
FAQ's:
View our frequently asked questions for more information.
Sample requirements:
We require 1-2 µg of genomic DNA (gDNA) with an OD260/280 ratio between 1.8 and 2.0, at a minimum concentration of 50 ng/μl, and a minimum volume of 20 μl. Higher concentrations are acceptable and will be diluted accordingly. click here.
Contact us today and one of our skilled account managers will be in touch with a free consultation including further information and pricing details.
Next Generation Sequencing
Source BioScience is one of Europe’s leading providers of commercial sequencing, offering Next Generation Sequencing services from our ISO accredited laboratories. We offer NGS services on the most prominent platforms including Illumina’s NovaSeq, NextSeq and MiSeq.